Day 11 of NTRESS 6000
Written on October 1, 2024
today we were working on data import, export, and conversion between data types!
first things first, we need to load tidyverse
library(tidyverse)
coronavirus <- read_csv('https://raw.githubusercontent.com/RamiKrispin/coronavirus/master/csv/coronavirus.csv')we have been working with this coronavirus dataset from here
coronavirus |>
filter(cases > 0) |>
group_by(date,type) |>
summarise(cases=sum(cases)) |>
ggplot() +
geom_col(aes(date,cases,fill =type))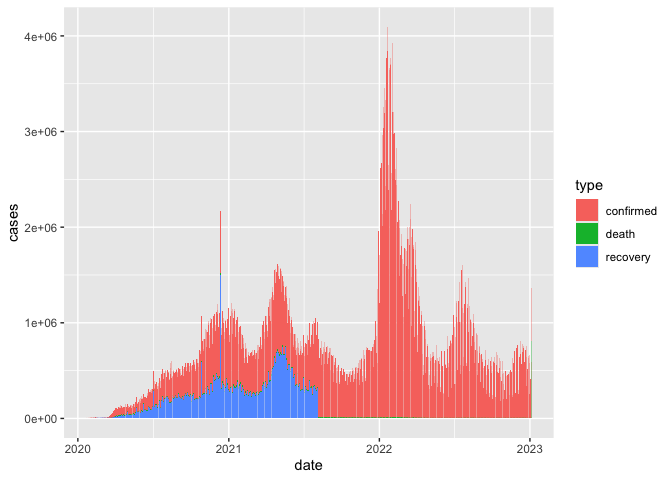
this is what our data looks like
## # A tibble: 6 × 15
## date province country lat long type cases uid iso2 iso3 code3
## <date> <chr> <chr> <dbl> <dbl> <chr> <dbl> <dbl> <chr> <chr> <dbl>
## 1 2020-01-22 Alberta Canada 53.9 -117. confirm… 0 12401 CA CAN 124
## 2 2020-01-23 Alberta Canada 53.9 -117. confirm… 0 12401 CA CAN 124
## 3 2020-01-24 Alberta Canada 53.9 -117. confirm… 0 12401 CA CAN 124
## 4 2020-01-25 Alberta Canada 53.9 -117. confirm… 0 12401 CA CAN 124
## 5 2020-01-26 Alberta Canada 53.9 -117. confirm… 0 12401 CA CAN 124
## 6 2020-01-27 Alberta Canada 53.9 -117. confirm… 0 12401 CA CAN 124
## # ℹ 4 more variables: combined_key <chr>, population <dbl>,
## # continent_name <chr>, continent_code <chr>
**lets filter this raw dataset to keep only the
date,
country,province,type,and
cases
we then are piping that dataframe into filter to
only keep cases on january 3rd, 2021
## # A tibble: 852 × 5
## date country province type cases
## <date> <chr> <chr> <chr> <dbl>
## 1 2021-01-03 Canada Alberta confirmed 3800
## 2 2021-01-03 United Kingdom Anguilla confirmed 2
## 3 2021-01-03 China Anhui confirmed 0
## 4 2021-01-03 Netherlands Aruba confirmed 32
## 5 2021-01-03 Australia Australian Capital Territory confirmed 0
## 6 2021-01-03 China Beijing confirmed 2
## 7 2021-01-03 United Kingdom Bermuda confirmed 0
## 8 2021-01-03 Netherlands Bonaire, Sint Eustatius and Saba confirmed 0
## 9 2021-01-03 Canada British Columbia confirmed 0
## 10 2021-01-03 United Kingdom British Virgin Islands confirmed 0
## # ℹ 842 more rowsokay i got too lazy with marking up this file so deal with some dirty code below!!
library(googlesheets4) #install.packages("googlesheets4")
gs4_deauth()
lotr_gs <- read_sheet("https://docs.google.com/spreadsheets/d/1X98JobRtA3JGBFacs_JSjiX-4DPQ0vZYtNl_ozqF6IE/edit#gid=754443596")#will help turn whatever you have into a number - helpful if your data isnt read as numeric
parse_double("1,23", locale = locale(decimal_mark = ","))## [1] 1.23## # A tibble: 6 × 5
## student_id full_name favourite.food mealPlan AGE
## <dbl> <chr> <chr> <chr> <chr>
## 1 1 Sunil Huffmann Strawberry yoghurt Lunch only 4
## 2 2 Barclay Lynn French fries Lunch only 5
## 3 3 Jayendra Lyne <NA> Breakfast and lunch 7
## 4 4 Leon Rossini Anchovies Lunch only <NA>
## 5 5 Chidiegwu Dunkel Pizza Breakfast and lunch five
## 6 6 Güvenç Attila Ice cream Lunch only 6library(janitor) #install.packages("janitor")
students |>
clean_names(case="lower_upper") |>
mutate(
age = parse_number(ifelse(age == "five", 5, age)))## # A tibble: 6 × 5
## studentID fullNAME favouriteFOOD mealPLAN age
## <dbl> <chr> <chr> <chr> <dbl>
## 1 1 Sunil Huffmann Strawberry yoghurt Lunch only 4
## 2 2 Barclay Lynn French fries Lunch only 5
## 3 3 Jayendra Lyne <NA> Breakfast and lunch 7
## 4 4 Leon Rossini Anchovies Lunch only NA
## 5 5 Chidiegwu Dunkel Pizza Breakfast and lunch 5
## 6 6 Güvenç Attila Ice cream Lunch only 6